Difference between revisions of "Using LiPD files"
Michaelerb (Talk | contribs) m (→A Note for Windows Users) |
Michaelerb (Talk | contribs) m (→LiPD Utilities in Python 3) |
||
| Line 75: | Line 75: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| − | From here, explore a LiPD file some more on your own. There are other commands in the LiPD utilities, but the ones above are enough to access the data on a basic level. If you'd like to use pre-built functions to explore the paleo data, see the Pyleoclim section farther down this page. | + | From here, explore a LiPD file some more on your own. There are other commands in the LiPD utilities, but the ones above are enough to access the data on a basic level. To get started with some more commands, read the quickstart guide on Github: [https://github.com/nickmckay/LiPD-utilities/blob/master/Examples/Welcome%20LiPD%20-%20Quickstart.ipynb Quickstart Guide]. |
| + | |||
| + | If you'd like to use pre-built functions to explore the paleo data, see the Pyleoclim section farther down this page. | ||
==Pyleoclim== | ==Pyleoclim== | ||
Revision as of 17:00, 27 June 2017
After downloading a LiPD file, there are a number of ways to use it. The recommended ways are to use the LiPD utilities or pyleoclim.
Contents
A Note for Windows Users
The LiPD utilities are compatible with Windows. However, most of the more advanced data analysis tools are only available for Mac or Linux platforms. If you're using Windows and would like to start using Linux, here are two options:
- Ask your university for an account on their Linux machine. Then use an ssh program like PuTTY to connect from your PC. An X-windows program like Xming is also needed to produce graphical windows.
- If option one isn't a possibility, you could install an Ubuntu virtual machine (e.g. VirtualBox) on your PC.
If you insist on working within Windows, you can install Python on your PC directly from the Python website. Make sure to select the Python 3 version.
After installing python, open a Windows command prompt. Despite cosmetic similarities, a Windows command prompt is different from a Linux terminal and generally uses different commands. To launch Python from here, type "python". To install python packages in Windows, exit python and type:
python -m pip install package_name
where "package_name" is the name of a python package, such as LiPD. However, not all packages may be available for Windows, so Linux is still recommended.
Another option is to use the Anaconda Python release.
LiPD Utilities
The LiPD Utilities are a primary way to interact with LiPD files. The utilities are available on GitHub in Matlab, R, and Python language. All three languages support reading and writing a LiPD file, extracting and collapsing time series, and filtering time series.
LiPD Utilities in Python 3
To use LiPD Utilies in Python, first make sure you have Python 3 installed. If you don't, one option is Anaconda.
Next install LiPD Utilities:
pip install LiPD
Start python with the command "python". Then import lipd:
import lipdThere are many functions in the LiPD utilities, but here are just a few. First, read a LiPD file:
lipd.readLipd('/path/to/data/file.lpd') # Load a specific LiPD file. Or...
lipd.readLipd() # Load a LiPD file through a GUI.After reading a LiPD file, use the extractTs() function to extract the data and metadata within the file:
data_all = lipd.extractTs()A list of dictionaries is now contained in the variable "data_all". (If you're unfamiliar with python, read a primer to get a better understanding.) Each dictionary contains data and metadata fields. From here, it's easy to start using the data:
data = data_all[0] # Save the first time series object to a new variable. Replace "0" with a different
# number to select a different time series object if there are more than one.
print(data.keys()) # Print the names of all data and metadata fields.
print(data['dataSetName']) # Print the contents of one field: the name of the data set.
year = data['year'] # Save the time values to a new variable.
values = data['paleoData_values'] # Save the data values to a new variable.Different data sets may not have all of the same fields, so use the ".keys()" command to check. Now, let's make a simple figure:
# Make a simple figure with matplotlib.
import matplotlib.pyplot as plt
plt.plot(year,values)
plt.title("Name: "+data['dataSetName']+", archive: "+data['archiveType'])
plt.xlabel(data['yearUnits'])
plt.ylabel(data['paleoData_variableName'])
plt.show()From here, explore a LiPD file some more on your own. There are other commands in the LiPD utilities, but the ones above are enough to access the data on a basic level. To get started with some more commands, read the quickstart guide on Github: Quickstart Guide.
If you'd like to use pre-built functions to explore the paleo data, see the Pyleoclim section farther down this page.
Pyleoclim
Pyleoclim is another primary way of interacting with LiPD files. While the LiPD Utilities offer a more manual approach to analysis, Pyleoclim has a variety of pre-built functions which can save time and effort. Install pyleoclim with:
pip install pyleoclim
To get started with pyleoclim, see the Pyleoclim wiki page. In particular, read the quickstart guide: Quickstart Guide.
Please note that some of the dependencies used by Pyleoclim are unavailable to Windows users.
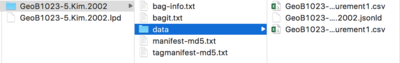
CSV files
If you need a plaintext version of the data, all LiPD files contain .csv files of the raw data. Simply unzip your LiPD file to find a .csv file. However, a central goal of LiPD is to put paleoclimate data into a standardized format which common analysis scripts can be built for, so using .csv files more than necessary is not recommended. Also, re-zipping an unzipped LiPD file will not turn it into a valid LiPD file.
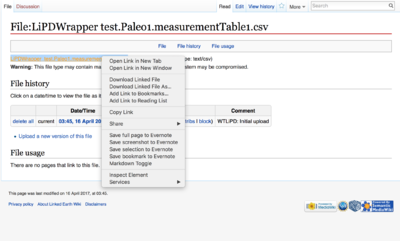
You can also access the .csv file on the wiki and download them.